About
Biography
The long-term goal of the Salomonis Lab is to develop broadly reusable immune modulatory therapies to target mis-splicing. Our basic premise is that independent genetic drivers of malignancy rely on common splicing alterations that represent novel targets for therapy and disease prevention. The role of splicing dysregulation in disease is profound, with >50% genetic diseases attributed to splicing factor mutations or transcript mis-splicing.
My research addresses major questions related to how splicing alterations occur, in which cell states, their impact on lineage specification, and recurrence across malignancies. To address these challenges, my lab has developed highly used bioinformatics approaches to resolve splicing impacts in disease, integrate diverse single-cell modalities, and redefine disease subtypes. This work focuses on approaches to exploit cutting-edge multi-omic techniques, deep learning and machine learning. Over the last decade, we have used these approaches to reveal novel isoforms regulating stem cell differentiation, splicing factor-mediated disease mechanisms, new disease subtypes and metastable transitional cell states.
I participate in multiple cell atlas initiatives, including the HCA, HuBMAP and LungMAP. Our current work applies deep learning to design new cancer vaccines, create advanced interactive cell atlases, identify novel isoforms that alter tumor extracellular signaling and game theory to resolve clonal heterogeneity in cancer.
BS: University of California, Los Angeles, CA, 1998.
PhD: University of California, San Francisco, CA, 2008.
Postdoctoral Fellow: Gladstone Institutes, San Francisco, CA, 2012.
Services and Specialties
Interests
Bioinformatics; genomics; cancer genomics; single-cell RNA-Seq analysis; alternative splicing; pathway analysis; pathway visualization; pathway curation; SIDS; stem cell biology; cardiac specification; renal graft dysfunction
Publications
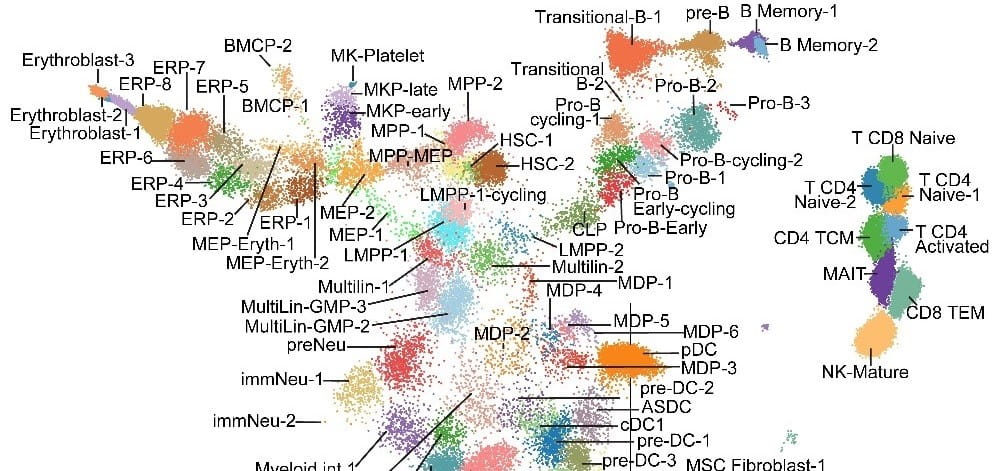
An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors. Nature Immunology. 2024; 25:703-715.
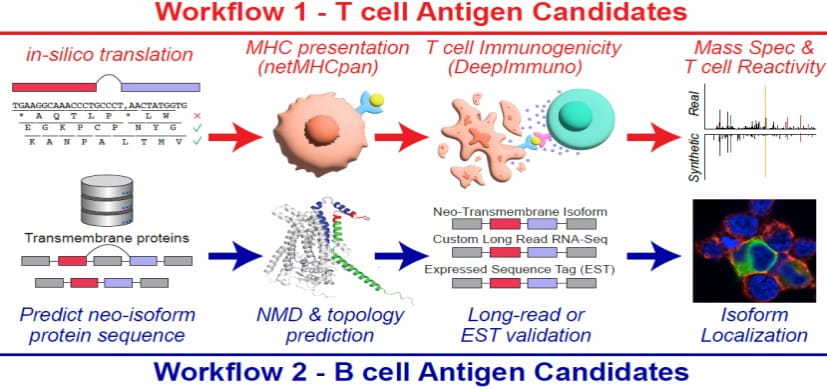
Splicing neoantigen discovery with SNAF reveals shared targets for cancer immunotherapy. Science Translational Medicine. 2024; 16:eade2886.
Decision level integration of unimodal and multimodal single cell data with scTriangulate. Nature Communications. 2023; 14:406.
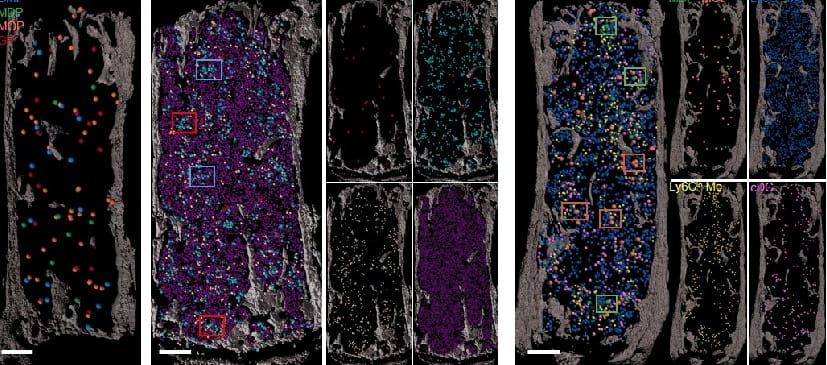
Erythroblastic islands foster granulopoiesis in parallel to terminal erythropoiesis. Blood. 2022; 140:1621-1634.
CellDrift: inferring perturbation responses in temporally sampled single-cell data. Briefings in Bioinformatics. 2022; 23:bbac324.
Bromodomain inhibition overcomes treatment resistance in distinct molecular subtypes of melanoma. Proceedings of the National Academy of Sciences of USA. 2022; 119:e2206824119.
A census of the lung: CellCards from LungMAP. Developmental Cell. 2022; 57:112-145.e2.
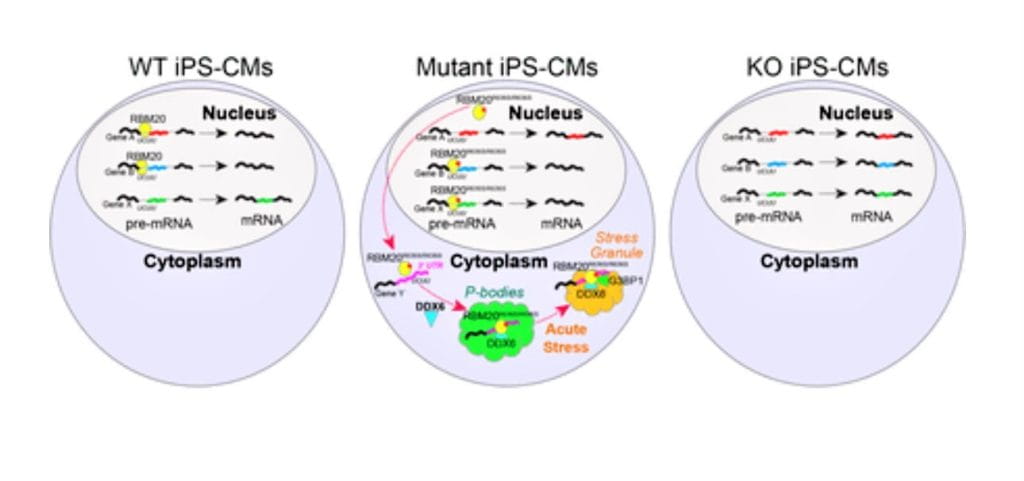
Gain-of-function cardiomyopathic mutations in RBM20 rewire splicing regulation and re-distribute ribonucleoprotein granules within processing bodies. Nature Communications. 2021; 12:6324.
DeepImmuno: deep learning-empowered prediction and generation of immunogenic peptides for T-cell immunity. Briefings in Bioinformatics. 2021; 22:bbab160.
From the Blog
Advanced Map of Human Blood Stem Cells Could Guide Highly Targeted Leukemia Care
Nathan Salomonis, PhD, H. Leighton "Lee" Grimes, PhD3/21/2024
New Research Tool Seeks to Accelerate Hunt for Cancer Immunotherapy Targets
Nathan Salomonis, PhD1/17/2024
Heart Tissue in a Dish Reveals New Links Between Neurodegeneration and Heart Disease
Nathan Salomonis, PhD11/3/2021
DeepImmuno: a Tool for Creating Custom T-cell Vaccines
Nathan Salomonis, PhD, Surya Prasath, PhD5/19/2021
Single Cell Approach Reveals Impact of Disease-Causing Gene Mutations
Nathan Salomonis, PhD, H. Leighton "Lee" Grimes, PhD3/22/2021
Bone Marrow ‘Map’ Opens Path to Organoid-like Blood Stem Cell Production
Nathan Salomonis, PhD, Daniel Lucas, PhD ...2/10/2021