H. Leighton "Lee" Grimes, PhD
- Director, Cancer Pathology Program, Division of Experimental Hematology & Division of Pathology
- Co-Leader, Program in Hematologic Malignancies of Cincinnati Children's Hospital Medical Center, Cancer and Blood Diseases Institute
- Professor, UC Department of Pediatrics
About
Biography
Determining the difference between important and unimportant DNA changes in childhood diseases can be tedious and difficult. The Grimes lab works to understand how normal hematopoiesis is programmed, and how diseases like marrow failure and leukemia change the transcriptional programming.
Dr. Grimes has a broad background in hematopoiesis, molecular biology and molecular oncology, including mouse modeling of hematopoiesis, myelopoiesis, marrow failure syndromes including myelodysplastic syndromes (MDS) and severe congenital neutropenia (SCN) and acute myeloid leukemia (AML).
He received a PhD in molecular pathology and immunology studying gene regulation with Maureen Goodenow (then at University of Florida). He then joined Philip Tsichlis (then at Fox Chase Cancer Center) when that lab was cloning novel genes activated by Moloney murine leukemia virus insertion mutagenesis (e.g., Akt, Tpl2).
Dr. Grimes participated in the identification of the Growth factor independent-1 (Gfi1) transcription factor, its DNA binding specificity, named the “SNAG” transcription repressor domain, and genetically linked this domain to Gfi1-directed biology.
The Grimes lab continues to focus on transcriptional integration of normal and malignant hematopoiesis.
With University of Washington colleague Marshall Horwitz, Dr. Grimes identified humans with mutations in Gfi1 who display severe congenital neutropenia (SCN) and non-immune chronic idiopathic neutropenia of adults (NI-CINA).
The Grimes lab has established multiple mouse models of human disease, including acute myeloid leukemia (AML), and more recently SCN. Their work has spanned both small molecule and RNA therapeutics.
In a 2016 study published in Cancer Discovery, they proved that DNMT3A haploinsufficiency could facilitate AML genesis.
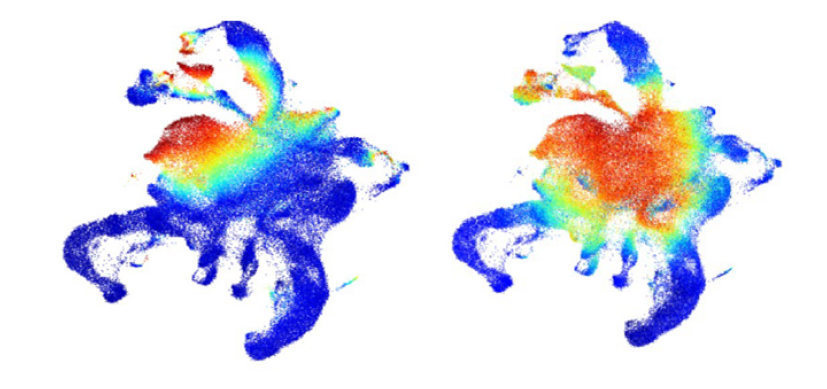
The Grimes lab was one of the first labs to utilize deep scRNA-seq profiling to dissect homeostatic myeloid development and provide deep molecular insight into the process of differentiation. This work was published in Nature in 2016.
They went on to generate the first mouse models of human SCN using patient-derived mutations in the Gfi1 transcription factor. This work was published in Nature in 2020.
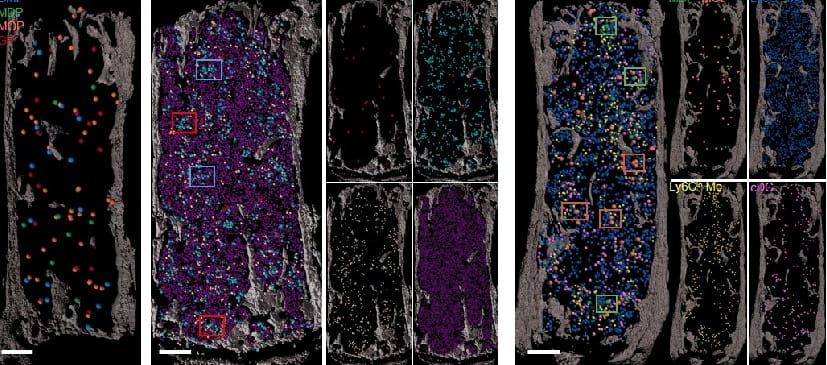
To determine the effects of SCN mutations, the team generated single-cell references for granulopoietic genomic states with linked epitopes, aligned mutant cells to their wild-type equivalents and identified differentially expressed genes and epigenetic loci. These insights facilitated the genetic rescue of granulocytic specification but not post-commitment defects in innate-immune effector function, and underscore the importance of evaluating the effects of mutations and therapy within each relevant cell state.
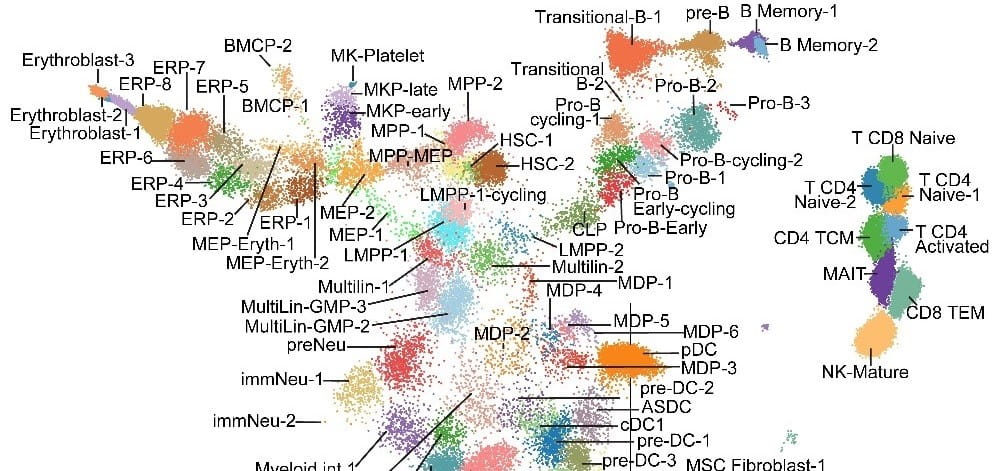
The Grimes lab is actively harnessing both established and cutting-edge single-cell technologies to dissect the transcriptional and epigenetic programming of normal and malignant hematopoiesis. In collaboration with Nathan Salomonis here at Cincinnati Children’s, they develop biologically-centric informatics algorithms to process single-cell data, web portals to disseminate the work flows, and web browsers to make the data easily accessible to biologists.
PhD: Immunology and Molecular Pathology, University of Florida, Gainesville, FL,
Fellowship: Postdoctoral, Fox Chase Cancer Center,
Services and Specialties
Interests
Acute myelogenous leukemia; T-cell acute lymphoblastic leukemia; severe congenital neutropenia; hematopoiesis; myelopoiesis; lineage decision; transcription factor
Research Areas
Publications
scTriangulate, a game-theory based framework for optimal solutions of uni- and multimodal single-cell data. bioRxiv. 2021; 2021.10.16.464640.
Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature. 2020; 582(7810):109-114.
Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016; 537(7622):698-702.
Activation of a branched-chain amino acid rheostat restores replication-dependent hematopoietic stem cell fitness. Cell Stem Cell. 2026; 33(2):289-305.e6.
Metallothionein 1 mediates growth and survival of Dnmt3a;Npm1-mutant acute myeloid leukemia. Haematologica. 2026; 111(1):373-378.
A branched chain amino acid rheostat controls hematopoietic stem cell replicative lifespan. Blood. 2025; 146(Supplement 1):1381.
5-azacytidine treatment reshapes clonal structure and alters mutation-dependent splicing in MDS patients. Blood. 2025; 146(Supplement 1):2054-2054.
A unified multimodal single-cell framework reveals a discrete state model of hematopoiesis in mice. Nature Immunology. 2025; 26(11):2086-2099.
Comprehensive characterization of human bone marrow microenvironment shows age-related changes. Haematologica. 2025; 110(11):2844-2848.
2003 – MOLECULAR DISSECTION OF AGING CLARIFIES THE IMPACT OF MDS GENETICS AND THERAPY. Experimental Hematology. 2025; 151:104916.
From the Blog
Advanced Map of Human Blood Stem Cells Could Guide Highly Targeted Leukemia Care
H. Leighton Grimes, PhD, Nathan Salomonis, PhD3/21/2024
Single Cell Approach Reveals Impact of Disease-Causing Gene Mutations
H. Leighton Grimes, PhD, Nathan Salomonis, PhD3/22/2021
Bone Marrow ‘Map’ Opens Path to Organoid-like Blood Stem Cell Production
H. Leighton Grimes, PhD, Daniel Lucas, PhD ...2/10/2021
Perspective on Single-Cell Analyses of Genetic Disease Models
H. Leighton Grimes, PhD4/23/2020
Finding Genetic Ripple Effects in a Single-Cell Environment
H. Leighton Grimes, PhD, Nathan Salomonis, PhD4/22/2020