Grimes Research Lab
The Grimes Lab at Cincinnati Children's works on hematopoiesis, response to infection and molecular oncology, including mouse modeling of hematopoiesis, myelopoiesis, marrow failure syndromes including myelodysplastic syndromes (MDS) and severe congenital neutropenia (SCN) and acute myeloid leukemia (AML).
The end goal is to provide fundamental biological and molecular insight into these diseases, and to illustrate new avenues to therapeutic intervention.
Led by H. Leighton "Lee" Grimes, PhD, our team is working toward an integrated multi-omics understanding of normal and malignant hematopoiesis. Trainees in the lab gain both wet-bench-based deep biological understanding of hematopoiesis, and (through mentoring from Nathan Salomonis, PhD), dry-bench facility with bioinformatics analytic tools and their applications.
With Marshall Horwitz (from the University of Washington) we identified humans with mutations in GFI1, who display severe congenital neutropenia (SCN) and non-immune chronic idiopathic neutropenia of adults (NI-CINA).
Since then, we have established multiple mouse models of human disease, including AML, and more recently SCN.
Our work has spanned both small molecule and RNA therapeutics.
- In Cancer Discovery 2016, we proved that Dnmt3a haplo-insufficiency could facilitate AML genesis.
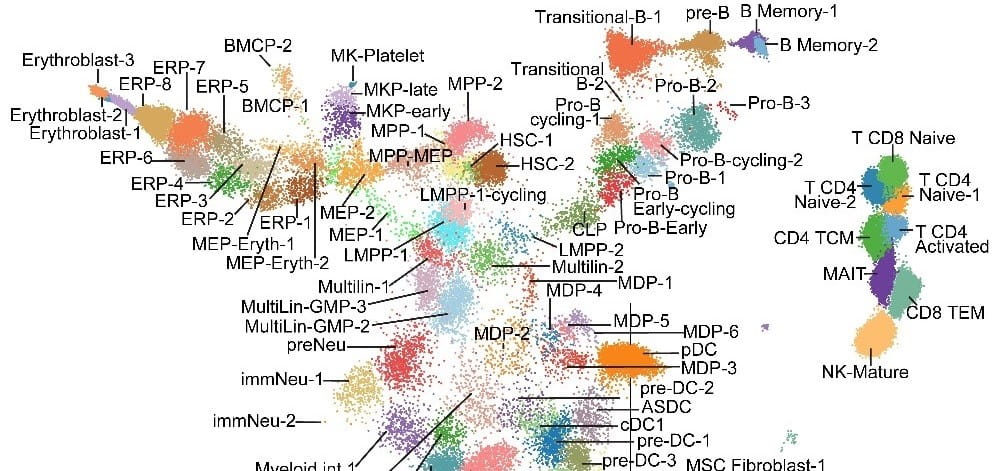
- In Nature 2016, we were one of the first labs to utilize scRNA Seq profiling to dissect homeostatic myeloid development and provide deep molecular insight into the process of differentiation.
- In Blood 2018, we showed (for the first time) that scRNA Seq data can be analyzed for RNA splicing, and showed that TGFbeta-induced destruction of SKI impacted splicing independent of splicing factor mutations in early-stage myelodysplasia (MDS).
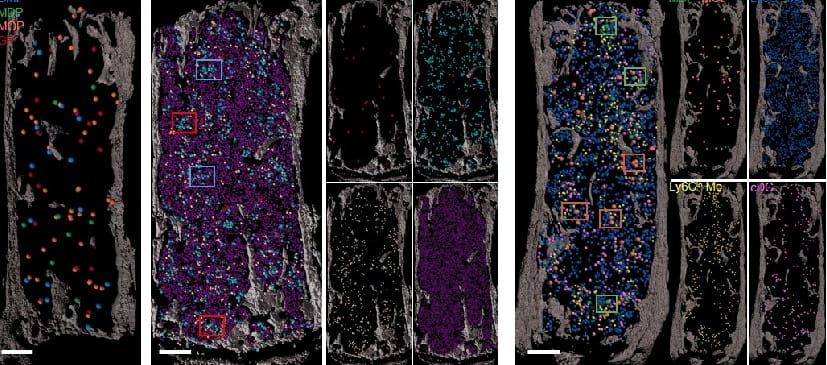
- In Nature June 2020, we generated the first mouse models of human severe congenital neutropenia (SCN) using patient-derived mutations in the GFI1 transcription factor. To determine the effects of SCN mutations, we generated single-cell references for granulopoietic genomic states with linked epitopes, aligned mutant cells to their wild-type equivalents and identified differentially expressed genes and epigenetic loci. These insights facilitated the genetic rescue of granulocytic specification but not post-commitment defects in innate-immune effector function and underscore the importance of evaluating the effects of mutations and therapy within each relevant cell state.
We are actively harnessing both established and cutting-edge single cell technologies to dissect the transcriptional and epigenetic programming of normal and malignant hematopoiesis.
Together with Salomonis Lab, we develop biologically-centric informatics algorithms to process single cell data, web portals to disseminate the work flows, and web browsers to make the data easily accessible to biologists. Interactive web viewers are available for our data analysis of human HSCP, and mouse neutrophil granulopoiesis.