Therapeutic Targeting of Mis-splicing in Cancer
The long-term goal of the Salomonis Lab is to develop broadly reusable immune modulatory therapies to target mis-splicing. Our basic premise is that independent genetic drivers of malignancy rely on common splicing alterations that represent novel targets for therapy and disease prevention.
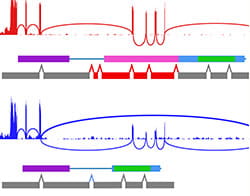
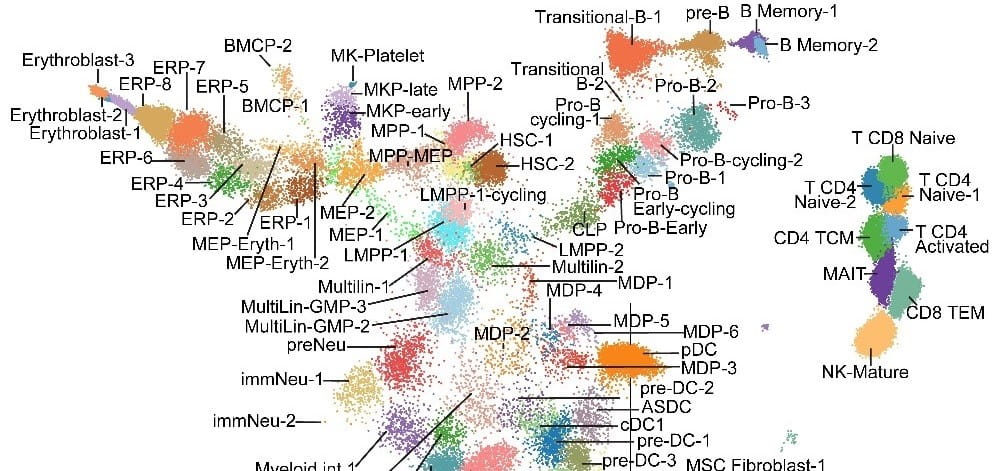
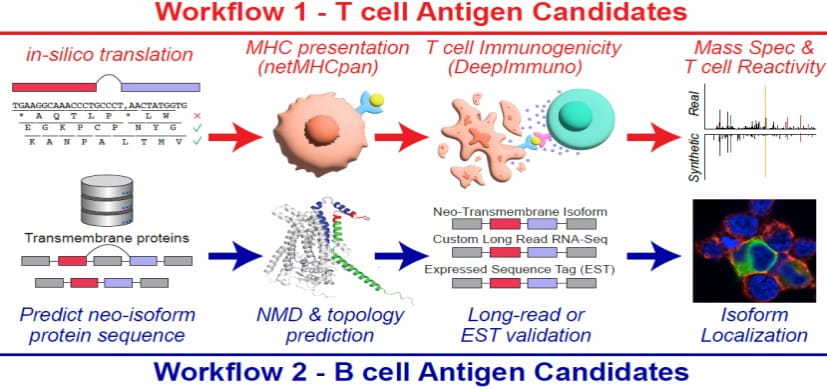
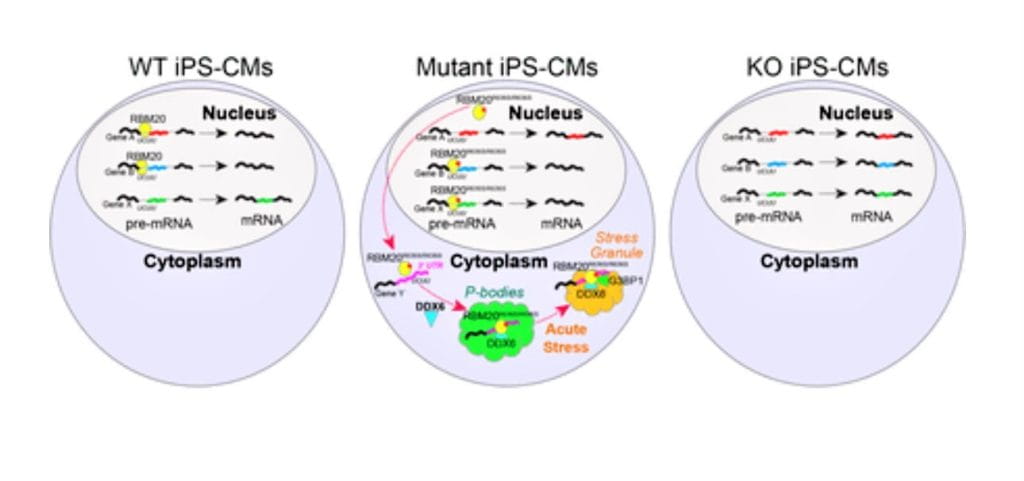
The role of splicing dysregulation in disease is profound, with >50% genetic diseases attributed to splicing factor mutations or transcript mis-splicing. Our lab’s research addresses major questions related to how splicing alterations occur, in which cell states, their impact on lineage specification, and recurrence across malignancies. To address these challenges, my lab has develops highly used bioinformatics approaches to resolve splicing impacts in disease, integrate diverse single-cell modalities, and redefine disease subtypes. These approaches include the creation of holistic workflows to develop therapeutic targets from alternative splicing (SNAF), predict immunogenic antigens (DeepImmuno), infer gene regulatory impacts from single-cell omics (scTriangulate, UDON, ICGS, cellHarmony) and define splicing subtypes and regulatory networks in cancer (OncoSplice, RNA-SPRINT). This work focuses on approaches to exploit cutting-edge multi-omic techniques, deep learning and machine learning. Over the last decade, we have used these approaches to reveal novel isoforms regulating stem cell differentiation, splicing factor-mediated disease mechanisms, new disease subtypes and metastable transitional cell states.
We aim to train the next generation of scientists interested developing new AI-driven computational biological approaches and apply such methods to answer key challenges in regulatory genomics. We welcome applications from potential graduate students and postdoctoral fellows who wish to drive innovation in these areas.