Hee Woong Lim, PhD
- Assistant Professor, UC Department of Pediatrics
- UC Department of Biomedical Informatics
About
Biography
I trained to be a computational scientist. However, after obtaining my PhD, I had a fascination with the convoluted and multifaceted, yet very ordered, biological system. Therefore, I moved my career in the direction of the bioinformatics field for my postdoctoral training at the University of Pennsylvania.
During my postdoc, I studied transcriptional regulations in multiple metabolic systems, including liver, adipose tissues and pancreas. Then, I joined the team in the Division of Biomedical Informatics at the Cincinnati Children’s Hospital Medical Center as an assistant professor and started a regulatory genomics laboratory to further my research.
In my research lab, I study fundamental mechanisms of gene transcriptional regulations in various contexts, such as metabolism, general cellular or tissue development, pathogenesis, and pharmacogenomics. In particular, I focus on enhancer regulations to delineate their intrinsic heterogeneity of architectures and functions using a high-resolution landscape of transcription factor binding and enhancer RNA (eRNA). To this end, my lab actively utilizes multi-omics high-throughput data including GRO-seq, RNA-seq, ChIP-seq, ChIP-exo, CUT&RUN, csRNA-seq, etc.
The most notable discoveries I made over the years include using eRNAs for direct monitoring of enhancer activities to uncover a molecular mechanism of anti-diabetic rosiglitazone-driven gene transcriptional regulation and an unconventional role of HDAC3 to prepare a thermogenic program in brown fat. In addition, I have discovered distinct binding modes (dimeric and monomeric binding) of the glucocorticoid receptor (GR) by studying the high-resolution footprints of GR binding using ChIP-exo.
I have more than 10 years of experience in the biomedical informatics field and began working at Cincinnati Children’s Hospital Medical Center in 2019. My research has been published in respected journals, such as Nature, Cell, Genome Research, Genes and Development, Proceedings of the National Academy of Sciences of USA, and Genome Research.
BS: Seoul National University, Seoul, Korea,
MS: Seoul National University, Seoul, Korea,
PhD: Seoul National University, Seoul, Korea,
Post Doc: University of Pennsylvania, Philadelphia, PA,
Interests
Regulatory genomics; enhancer; transcription; metabolism; pharmacogenomics; machine learning
Research Areas
Publications
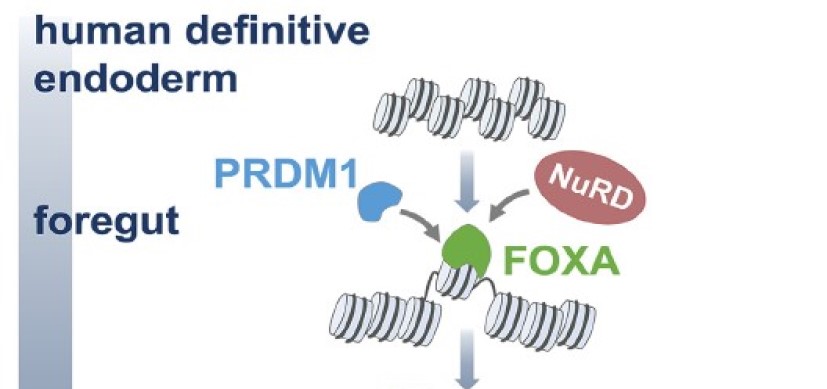
Pioneer and PRDM transcription factors coordinate bivalent epigenetic states to safeguard cell fate. Molecular Cell. 2024; 84(3):476-489.e10.
Single-cell RNA-Seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI Insight. 2022; 7(11).
Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2018; 115(22):E5096-E5105.
Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017; 546(7659):544-548.
Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Research. 2015; 25(6):836-844.
An inducible system to study the regulatory functions of GSX2 in human lateral ganglionic eminence-like progenitors. Developmental Biology. 2026; 532:35-51.
The Transcription Factor TCF21 is Necessary for Adoption of Cell Fates by Foxd1+ Stromal Progenitors during Kidney Development. American Journal of Physiology - Renal Physiology. 2026.
Defective Notch1 signaling in endothelial cells drives pathogenesis in a mouse model of Adams-Oliver syndrome. Journal of Clinical Investigation. 2025; 135(23).
PRDM16 regulates smooth muscle cell identity and atherosclerotic plaque composition. Nature Cardiovascular Research. 2025; 4(11):1573-1588.
Proximal Tubule Cells Contribute to the Thin Descending Limb of the Loop of Henle during Mouse Kidney Development. Journal of the American Society of Nephrology. 2025; 36(10):1928-1938.
From the Blog
How ‘Pioneers’ Blaze the One Trail that Determines Cell Fate
Hee Woong Lim, PhD, Makiko Iwafuchi, PhD1/10/2024