Leah C. Kottyan, PhD
- Research Director, Division of Allergy & Immunology
- Professor, UC Department of Pediatrics
About
Biography
I am a researcher in the Department of Eosinophilic Disorders and the Center for Autoimmune Genomics and Etiology, where my laboratory studies the genetic etiology of diseases that have an immunological component. I came to Cincinnati Children’s in 2005.
I’m particularly interested in the way genetic variants increase the risk for eosinophilic esophagitis, systemic lupus erythematosus, atopic dermatitis and multiple sclerosis. Because many of the genetic risk variants do not change the protein amino acid code, a lot of our research is focused on genotype-dependent transcriptional regulation in disease-relevant cells and tissues.
I became interested in these areas after leading many genetic studies aimed at identifying polymorphisms in the genome that increase disease risk. As an immunologist and molecular biologist, I want to understand what molecular mechanisms mediate a change in DNA sequence that leads to genotype-dependent cell biology in a way that increases disease risk.
One of the joys of my career is being a part of multidisciplinary teams in which my colleagues and I use our individual strengths and expertise to perform research together that none of us could do alone. In this way, we have had the privilege to make numerous notable discoveries.
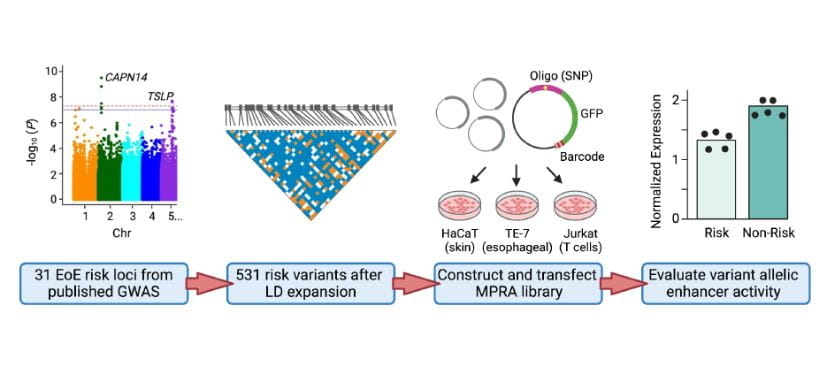
For example, I’ve been part of teams that identified the genetic variations near the esophageal-specific gene CAPN14 that increase the risk for eosinophilic esophagitis. I was also part of a team that identified the interaction of transcriptional regulators of the Epstein Barr virus with autoimmune disease risk variants. On another team, we identified estrogen receptor's genotype-dependent interaction with genetic variants that increase risk for pre-term birth.
I share a wet lab with Dr. Matthew Weirauch at Cincinnati Children’s. The Weirauch lab develops new computational tools to understand transcriptional regulation. A major strength of this arrangement is the mixing of experimental and computational backgrounds among topics, and therefore trainees.
The multidisciplinary structure among the principle investigators and their teams provides the conceptual synergy driving many collaborative projects. Dynamic interactions between people performing functional genomic experiments and the people developing the analytical pipelines in this lab enables high-quality, robust scientific discovery. We have made incredible progress over the past few years toward identifying the transcriptional mechanisms that drive genotype-dependent gene expression and disease risk.
During my career, I’m proud to have received the Cincinnati Children’s 2020 Mentoring Achievement Award, the 2018 Recognition of Excellence Award, and the 2016 Darrell Goll Award. The Darrell Goll Award recognizes the most outstanding new investigator publishing in the calpain field over the past three years. I am also pleased to serve as a coordinator for the Schmidlapp Young Women Scholars Program, which provides career support to women undergraduate scholars at Cincinnati Children’s.
BA: Chemistry and Cell Biology, Huntingdon College, Montgomery, AL, 2005
PhD: Immunobiology, University of Cincinnati, Cincinnati, OH, 2010
Services and Specialties
Interests
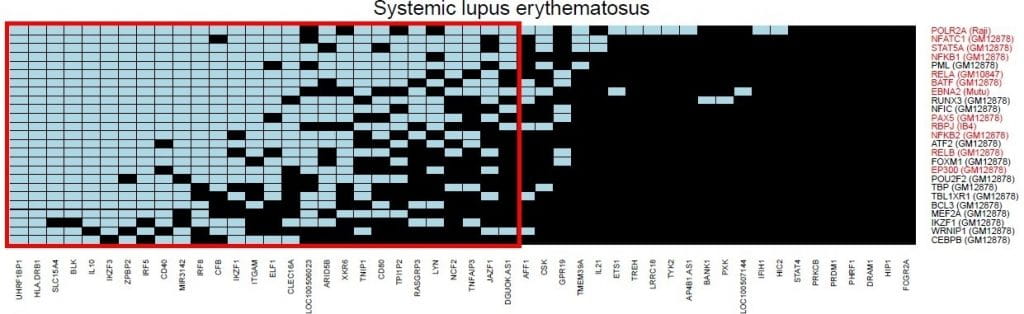
Genetic basis of lupus; genetic basis of eosinophilic esophagitis; immunological mechanisms mediating genetic association with disease
Publications
Update on the genetics of allergic diseases. Journal of Allergy and Clinical Immunology. 2025; 155(6):1738-1752.
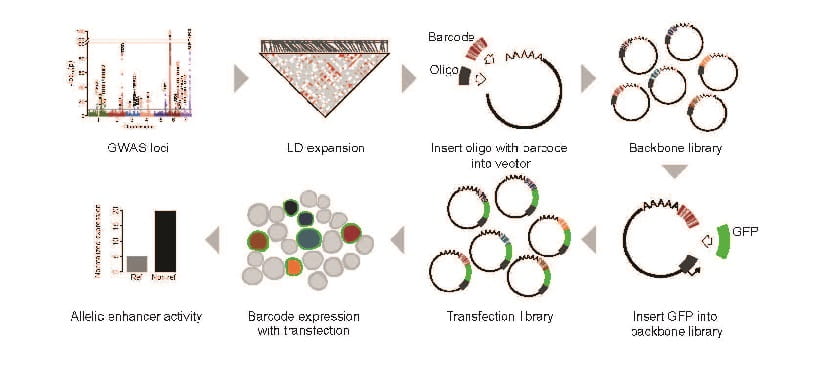
Massively Parallel Analysis of Genotype-Dependent Enhancer Activity Among Atopic Dermatitis Genetic Risk Variants. . 2024.
Shared and distinct interactions of type 1 and type 2 Epstein-Barr Nuclear Antigen 2 with the human genome. BMC Genomics. 2024; 25(1):273.
Systematic identification of genotype-dependent enhancer variants in eosinophilic esophagitis. American Journal of Human Genetics. 2024; 111(2):280-294.
Multiancestral polygenic risk score for pediatric asthma. Journal of Allergy and Clinical Immunology. 2022; 150(5):1086-1096.
Epigenetic and transcriptional dysregulation in CD4+ T cells in patients with atopic dermatitis. PLoS Genetics. 2022; 18(5):e1009973.
Epstein-Barr virus nuclear antigen 2 extensively rewires the human chromatin landscape at autoimmune risk loci. Genome Research. 2021; 31(12):2185-2198.
Runx1 shapes the chromatin landscape via a cascade of direct and indirect targets. PLoS Genetics. 2021; 17(6):e1009574.
Global discovery of lupus genetic risk variant allelic enhancer activity. Nature Communications. 2021; 12(1):1611.
From the Blog
National Academies Report Charts a Path to Strengthen NIH-Funded Pediatric Research
Leah C. Kottyan, PhD2/6/2026
Researchers Produce Annotated Molecular Map of Eosinophilic Esophagitis
Leah C. Kottyan, PhD, Matthew T. Weirauch, PhD ...1/26/2024
‘Massively Parallel’ Gene Screening Tool Can Accelerate Research for Nearly Any Disease
Leah C. Kottyan, PhD, Matthew T. Weirauch, PhD ...3/12/2021
‘Mono’ Virus Linked to Seven Serious Diseases
Leah C. Kottyan, PhD, Matthew T. Weirauch, PhD6/30/2019