Matthew T. Weirauch, PhD
- Member, Center for Autoimmune Genomics and Etiology, Divisions of Human Genetics, Biomedical Informatics, and Developmental Biology
- Professor, UC Department of Pediatrics
About
Biography
I am a bioinformatician geneticist who works to achieve a thorough understanding of human and viral transcriptional regulation mechanisms in complex diseases. Since joining Cincinnati Children’s in 2012, I’ve helped develop new computational tools and experimental data to attain a mechanistic understanding of gene regulation.
Our lab is highly collaborative; my colleagues and I work on a wide range of biological and disease topics. We also take a general approach of using all available data to generate and ultimately validate new hypotheses. We’ve long been interested in gene regulation because it’s a fundamental process underlying all life. It’s the first step in translating the “instructions” that are encoded in DNA into various functions. More recently, we’ve expanded our focus to include the role of viruses in human gene regulation and how this contributes to human disease processes.
My colleagues and I discovered (and continue to study) a critical role for the Epstein-Barr virus EBNA2 protein in the mechanistic processes underlying several autoimmune diseases. We are currently examining how the hundreds of regulatory proteins encoded by other viruses contribute to other human diseases. We’ve also constructed several widely used databases that summarize current knowledge about all eukaryotic and viral gene regulatory proteins.
I’ve received several professional honors and appointments, including:
- Cincinnati Children’s Hospital Research Achievement Award (2018)
- Elected to the Faculty of 1000 literature service, Systems and Network Biology Section (2018-present)
- Cincinnati Children’s Research Fund Endowed Scholar Award (2017-2020)
- Ranked 5th among the “Top Ten Papers in Regulatory and Systems Genomics in 2014” for Weirauch et al. Cell2014, as selected by participants of the 2014 RECOMB/ISCB Conference on Regulatory and Systems Genomics
Postdoctoral Fellow: University of Toronto (Donnelly Center for Cellular and Biomolecular Research),, Toronto, Ontario, Canada,
PhD: Bioinformatics, University of California Santa Cruz, Santa Cruz, California,
BSc: Computer Science, Pennsylvania State University, University Park, PA,
Services and Specialties
Interests
Gene regulation Bioinformatics Functional Genomics
Publications
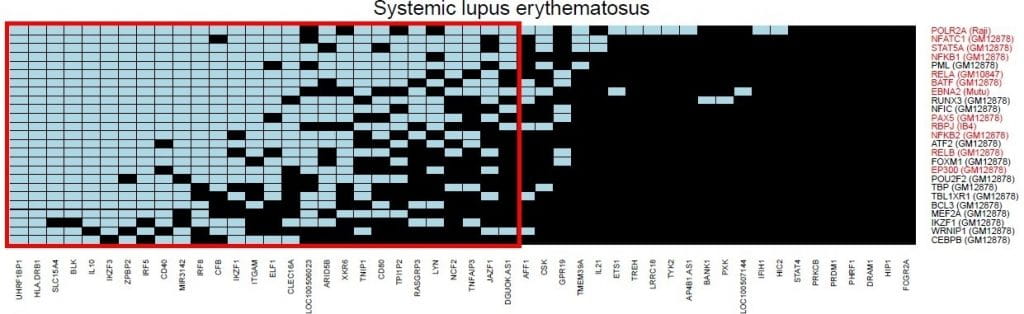
Global discovery of lupus genetic risk variant allelic enhancer activity. Nature Communications. 2021; 12(1):1611.
Global discovery of lupus genetic risk variant allelic enhancer activity. Nature Communications. 2021; 12(1).
Human Virus Transcriptional Regulators. Cell. 2020; 182(1):24-37.
Similarity regression predicts evolution of transcription factor sequence specificity. Nature Genetics. 2019; 51(6):981-989.
Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nature Genetics. 2018; 50(5):699-707.
The Human Transcription Factors. Cell. 2018; 172(4):650-665.
Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. New England Journal of Medicine. 2017; 377(12):1156-1167.
Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nature Biotechnology. 2015; 33(8):831-838.
Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158(6):1431-1443.
A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499(7457):172-177.
From the Blog
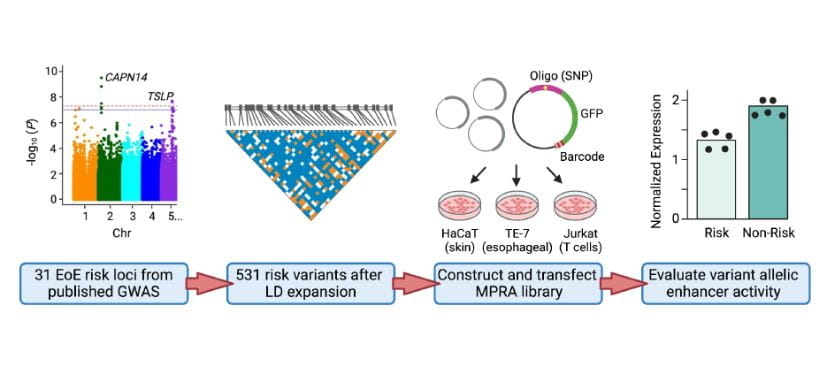
Researchers Produce Annotated Molecular Map of Eosinophilic Esophagitis
Matthew T. Weirauch, PhD, Leah C. Kottyan, PhD ...1/26/2024
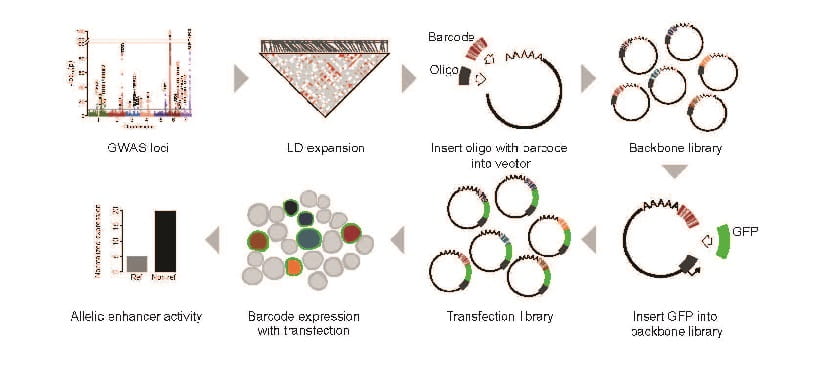
‘Massively Parallel’ Gene Screening Tool Can Accelerate Research for Nearly Any Disease
Matthew T. Weirauch, PhD, Leah C. Kottyan, PhD ...3/12/2021
‘Mono’ Virus Linked to Seven Serious Diseases
Matthew T. Weirauch, PhD, Leah C. Kottyan, PhD6/30/2019