Kottyan Research Lab
Overall focus: The Kottyan Lab is focused on identifying the molecular mechanisms that underlie specific genetic associations with human disease.
Why do some individuals develop disease? Broadly speaking, genetic risk polymorphisms and environmental exposures act in concert with stochastic factors to influence risk for many diseases. Recent studies are beginning to identify the disease mechanisms impacted by specific genetic risk variants and environmental exposures. In particular, functional genomics-based approaches are revealing that changes in human chromatin state and gene expression are important molecular mechanisms through which genetic and environmental factors influence human disease risk. Understanding disease mechanisms is vitally important for the development of treatments and therapies. In the past two decades, thousands of genetic variants with roles in >1,500 human diseases have been identified. Importantly, genetic association studies do not identify causal SNPs, critical genes, or etiological pathways. There is a pressing need for new approaches that translate the massive results from genetic studies into clear mechanistic understandings of the genetic and environmental contributions to human diseases.
Drs. Leah Kottyan and Matthew Weirauch have brought together their wet laboratories to form a shared Transcription Factor Genetics working group. A major strength of this arrangement is the mixing of experimental and computational backgrounds among topics, and therefore trainees. The multidisciplinary complementation among the principle investigators and their teams provides the conceptual synergy driving many collaborative projects. Dynamic interactions between people in the wet lab performing functional genomic experiments and the people developing the analytical pipelines enables high quality, robust scientific discovery. Along with our division director, Dr. John Harley, we have made incredible progress over the past few years towards the identification of transcriptional mechanisms that drive genotype-dependent gene expression and disease risk.
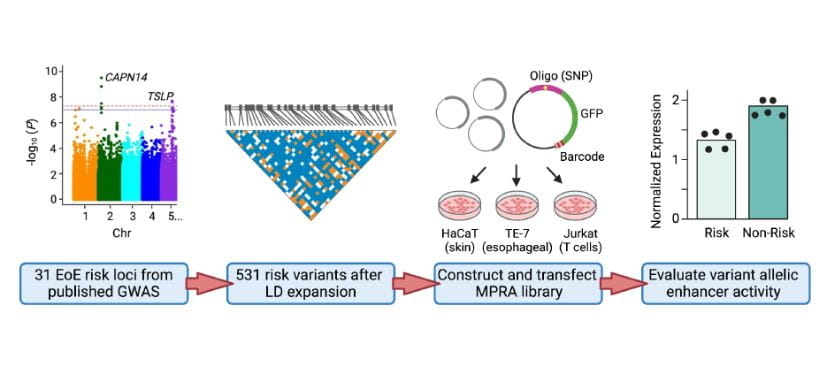
Genome-wide association studies (GWAS) made it financially and logistically possible to use tens of thousands of people with and without the disease to identify pieces of DNA that increase disease risk and, equally importantly, to confirm these associations in independent replication cohorts. The National Institutes of Health and private research organizations poured hundreds of millions of dollars into these studies, and we now have long lists of DNA loci that are associated with diseases such as systemic lupus erythematosus, multiple sclerosis, and atopic dermatitis. Dr. Kottyan led one such study for eosinophilic esophagitis (EoE) and identified 9 new risk loci including a locus encoding CAPN14, a gene that is specifically upregulated as a function of disease activity and genetic haplotype.
In our genetic analyses, we start with genetic data and apply rigorous frequentist and Bayesian statistical analysis using open source analytical tools. Then, we use in silico prediction tools to identify testable allelic hypothesis (i.e. we design experiments that compare the biological consequences of two alleles of a genetic variant). Finally, we perform experiments with the aim of identifying possible biological mechanisms that can explain the genetic associations identified in the initial studies.
Every field of science approaches scientific inquiry from a different orientation. In genetics, the big data and complex statistical analyses are intoxicating. For the past decade, many geneticists have approached genetic analysis with the idea that we just need to increase statistical power by enlarging the sample size (e.g. ~100,000 person GWAS) and assess enough loci (e.g. the OMNI5 with 5 million markers that can be imputed to over 12 million variants) in order to find the “genes” associated with a disease. Unfortunately, clinicians can do relatively little with the correlative genetic information currently available for most autoimmune diseases (e.g. p-values and odds ratios). Instead of sulking about the limited “success” of the GWAS era, our lab celebrates the identification of these loci in the human genome that increase disease risk and uses them to understand the way that single genetic variants can change molecular and cellular activity in a manner that leads to disease.