Emily R. Miraldi, PhD
- Member, Division of Immunobiology
- Member, Division of Biomedical Informatics
- Associate Professor, UC Department of Pediatrics
About
Biography
Our research goal is immuno-engineering: to alter the behavior of specific immune cell populations in disease contexts (autoimmune disease, organ transplant, and cancer) without compromising the body's homeostatic immune function (e.g. defense against pathogens). Thus, it is critical to develop a nuanced understanding of how different immune cells sense and respond to environmental cues across the body, in both physiological and disease settings. To this end, our lab's major focus is reverse-engineering the underlying logic of immune cells (molecular networks that drive cellular responses) from high-dimensional molecular measurements of immune cells in action (sensing and responding to perturbations, disease conditions, etc.).
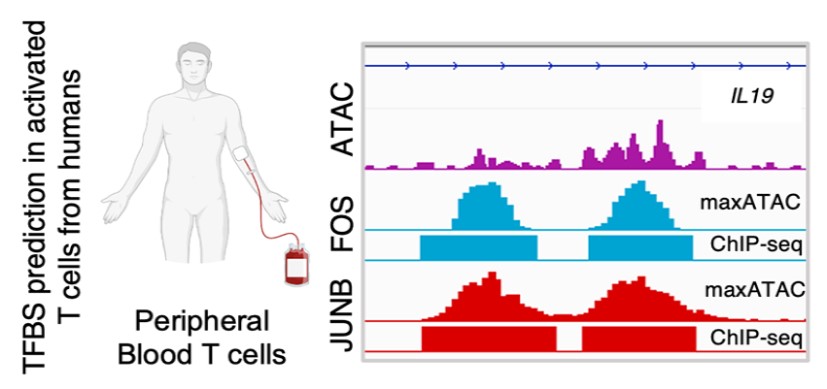
The lab's focus is transcriptional regulatory network inference, modeling gene expression as a function of transcription factor activities, from gene expression and measurements of chromatin state. Chromatin accessibility measurements by ATAC-seq, together with transcription-factor DNA-binding preferences (motifs), can be used to broadly profile potential transcription factor binding events in relatively small populations of cells. Thus, we have used ATAC-seq with RNA-seq to enable de novo inference of transcriptional regulatory networks in physiological settings where sample material is limiting (e.g., intestinal immune cells in response to microbial/genetic perturbations). To date, most of these efforts have been in mouse models, as it is generally not possible to obtain sufficient sample material for similar experimental designs in human. To build human immune cell models, we are developing multi-task learning approaches to leverage evolutionarily conserved relationships and borrow statistical power from mouse datasets for inference in human. We are also developing methods to enable transcriptional regulatory network inference in very rare cell populations, as measured from single-cell RNA-seq experiments.
Interests
Systems biology; computational immunology; immuno-engineering; network inference
Research Areas
Publications
maxATAC: Genome-scale transcription-factor binding prediction from ATAC-seq with deep neural networks. PLoS Computational Biology. 2023; 19(1):e1010863.
Leveraging chromatin accessibility for transcriptional regulatory network inference in T Helper 17 Cells. Genome Research. 2019; 29(3):449-463.
Multi-study inference of regulatory networks for more accurate models of gene regulation. PLoS Computational Biology. 2019; 15(1):e1006591.
Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nature Immunology. 2017; 18(4):412-421.
4C-ker: A Method to Reproducibly Identify Genome-Wide Interactions Captured by 4C-Seq Experiments. PLoS Computational Biology. 2016; 12(3):e1004780.
Sparse and compositionally robust inference of microbial ecological networks. PLoS Computational Biology. 2015; 11(5):e1004226.
Molecular network analysis of phosphotyrosine and lipid metabolism in hepatic PTP1b deletion mice. Integrative Biology (United Kingdom). 2013; 5(7):940-963.
B-Cell Repertoire of Children With Atopic Dermatitis Exhibits Altered IgE Maturation Associated With Allergic Food Sensitization. Allergy. European Journal of Allergy and Clinical Immunology. 2026; 81(2):513-523.
Implications of Model Loss and Configuration for Sparse Histological Segmentation. In: Machine Learning Methods in Biomedical Field. Springer Nature; 2026:189-217.
From the Blog
New Tool Provides Low-Cost, High-Quality Means to Map Transcription Factor Binding Genome-Wide
Emily R. Miraldi, PhD4/5/2023
What is the Inferelator? How New Software Can Track Complex Transcription Factor Patterns
Emily R. Miraldi, PhD9/7/2022
‘Massively Parallel’ Gene Screening Tool Can Accelerate Research for Nearly Any Disease
Emily R. Miraldi, PhD, Leah C. Kottyan, PhD ...3/12/2021