Miraldi Research Lab
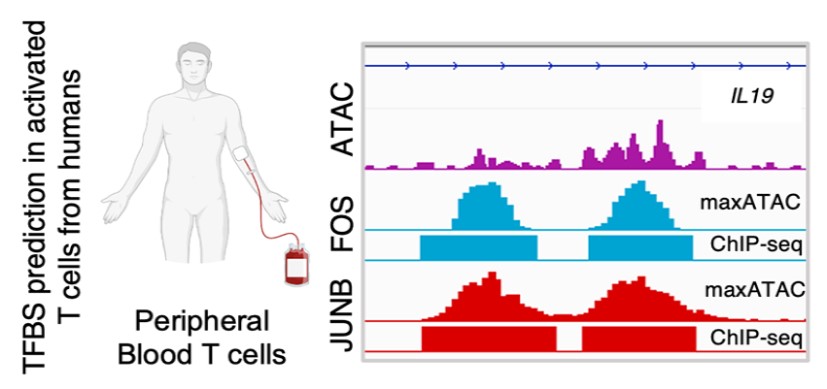
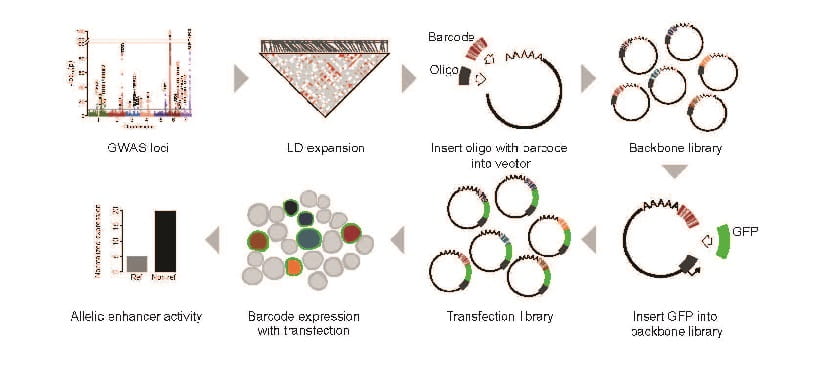
The Miraldi lab’s focus is mathematical modeling of the immune system from high-dimensional genomics measurements. In close collaboration with experimental immunologists, we seek to learn how diverse immune cells sense and respond to their environment in both health and disease. Our studies leverage new biotechnologies (e.g., chromatin accessibility, single-cell genomics measurements) and often require development of new computational methods. The resulting genome-scale models provide unbiased, experimentally testable hypotheses. Long-term, we would like to use these models to re-engineer immune-cell behavior in the context of autoimmunity, cancer and other diseases.
In the News
Deep neural networks to predict transcription factor binding, Ohio Supercomputer Center, Aug 24, 2023.
New tool provides low-cost, high-quality epigenome maps, Research Horizons, April 5, 2023.
Smart Software Untangles Gene Regulation in Cells, Nature, September 5, 2022.
Patient-Specific Prediction of Epigenomes Through Deep Learning, January 2021.
A New Way to Map Cell Regulatory Networks. EurekAlert!, March 5, 2019.
How Big Data and Genomics are Improving Infectious Disease Research. Healthcare Analytics, Nov. 30, 2017.
Immuno-Engineering from the Numbers. Research Horizons, Spring 2017.
Computational Methods and Biological Systems
Learn more about Miraldi Lab research projects.
Meet the Lab
Learn more about our current lab members and lab alumni.
Open Lab Positions
The Miraldi Lab seeks talented, motivated individuals at all levels (masters, PhD, postdoc, programmers). Interested parties are invited to apply today.