Proteomics driven biomarker and airway disease pathway discovery
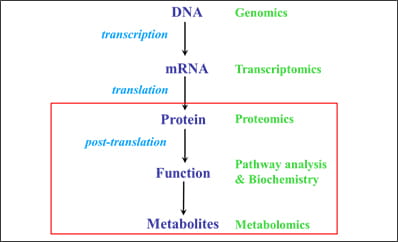
Our systems biology approach combines a number of methods to study the molecular basis of CF disease. For the program of biomarkers and disease pathways identification, we depend heavily on proteomic analyses. The lab uses state of the art software to study and analyze cellular pathways in conjunction with our proteomic studies using our dedicated LC mass spectrometer to gain insight into the systems biology of inflammatory lung disease, particularly for cystic fibrosis (CF).
We are working with the CF Foundation’s Biomarker Consortium to advance the biomarkers from clinical applications of diagnosing and prognosticating disease severity. We have several projects completed and some underway for the program of biomarkers and disease pathways identification: First, we examined early markers of lung disease that are specific for CF; second, we investigated surrogate markers for disease severity in CF; third, we sought to discover robust biomarkers in CF that could serve as diagnostic predictors of phenotype, or identify patients who may optimally respond to novel therapeutics.
Following the results of these studies, we investigated whether the identified inflammatory protein and lipid abnormalities associated with CF lung disease discriminate CF children with mild versus more severe underlying lung disease. In addition, we set out to determine the feasibility of utilizing banked serum samples from the CF Foundation Therapeutics (CFFT) Biorepository linked to clinical data in the CFF Patient Registry for CF biomarker development.
 |
|
Figure 1. Our approach combines a number of methods to study the molecular basis of CF disease |
CF lung disease begins in infancy, and is characterized by mucus plugging, infection with inflammation, air trapping, and structural lung changes (eg: bronchiectasis) that are readily detectable by three years of age. Early markers of lung disease that are specific for CF, however, are lacking. This information is critical to our understanding of what initiates CF lung disease, and to identify proximal targets for intervention. This lack of specificity may in part represent examination that has been limited to candidate-based analysis of biofluids, coupled with comparisons with inadequate disease controls. We hypothesized that using an unbiased and blinded approach, proteomic analysis of BALF from young CF patients without chronic Pseudomonas infection compared with non-CF disease controls (ages birth-5 years) would identify disease pathways and biomarkers associated with early CF lung disease.
Our results indicate that CF BALF contains protein signatures that demonstrate increased metabolism, increased catabolism and an alteration in pathways that regulate lipid, alcohol, and iron metabolism during infection. These significant differences in CF vs non CF infected controls suggests that the CF BALF signature can be segregated from non CF disease controls, exhibiting different biomarkers that delineate pathways that may be informative about early disease pathogenesis and also serve as potential therapeutic targets.
 |
|
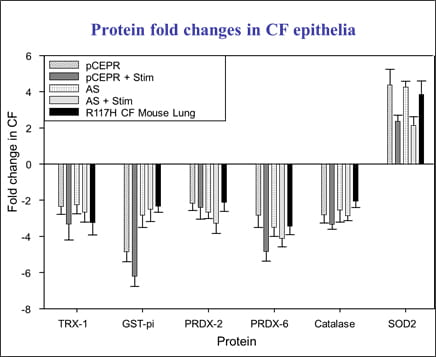
Figure 2. Protein fold changes in cultured CF epithelia and CF mouse whole lung. |
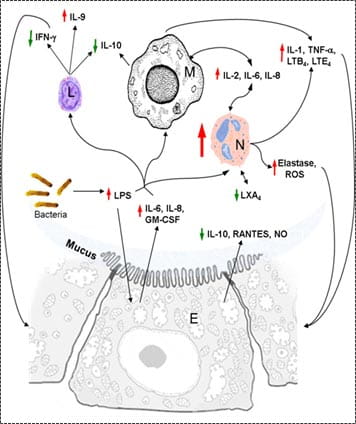 |
| Figure 3. Schematic of changes in protein levels on inflammation in CF. |
In addition, surrogate markers for disease severity in CF that can be rapidly and reliably measured to establish change in the course of the disease in response to therapy are similarly highly desirable. We proposed that changes in biological networks identified in blood between CF patients with severe versus mild disease reflect lung function status. For example, measurement of short term markers that track with or predict long term changes in FEV1 may shorten the length and significantly reduce the cost of clinical trials. As part of the CFF Biomarker Consortium’s serum protein biomarker discovery project, we analyzed differences in pathways in serum samples from 44 children with mild lung disease (FEV1>80th percentile) and 44 with more severe lung disease (FEV1<45th percentile). Deep proteomic analyses of serum samples from CF patients segregated by disease severity reveal the differential expression of protein biomarkers that cluster into pathways that regulate the differentiation of cartilage, myeloid leukocytes, and intestinal epithelia. Direct analysis of these pathways may provide a rapid estimation of changes in the severity of disease and may be a useful outcome measure of therapeutic efficacy.
Furthermore, robust biomarkers in CF could serve as diagnostic predictors of phenotype, or identify patients who may optimally respond to novel therapeutics. Serum proteomics by mass spectrometry offers identification and relative quantification of thousands of expressed protein isoforms representative of molecular processes; however, it presents the classic ‘big data’ problem. Our aim was to reduce an exploratory assemblage of 61,941 protein isoforms to a panel of markers that discriminate CF disease severity (to be further validated experimentally). Our method reduced massive dimensionality to a candidate panel of isoforms predictive of CF disease severity. Small sample size, skewed data, and the dominating effect of zero detection impede cross-validation techniques and limit statistical power using standard methods. Further refinement of this panel and proteomic pathway analysis will support transition from ‘discovery’ to ‘validation’ of protein markers in CF.
An important question is whether blood-based markers are sensitive enough to discriminate between lung disease severity, given that the inflammatory response in CF is largely confined to the lung. The purpose of this study was to investigate whether previously identified inflammatory protein and lipid abnormalities associated with CF lung disease discriminate CF children with mild versus more severe underlying lung disease. A secondary objective was to determine the feasibility of utilizing banked serum samples from the CF Foundation Therapeutics (CFFT) Biorepository linked to clinical data in the CFF Patient Registry for CF biomarker development. Several candidate serum inflammatory protein and lipid levels differ in CF children segregated by lung disease severity, further validating these as markers in CF. In addition, we have demonstrated the value of maintaining centralized, high quality patient derived samples for future research, with linkage to clinical information to answer testable hypotheses in biomarker development.



