Single Cell Applications in Lung Development and Diseases
The objective of our team in LungMAP2 project seeks to work together with other research centers and consortiums (HCA, HubMAP) to develop analytic pipelines to understand human lung development from the saccular, alveolar stages of morphogenesis at single cell level of resolution. We will integrate single cell transcriptomic and chromatin accessibility (ATAC-seq) to characterize cells and cell-cell interactions in human lung at both normal developmental and pediatric disease conditions.
- Lung development – identify cell types, signature genes, regulatory networks and cell-cell interactions at critical stages of lung development.
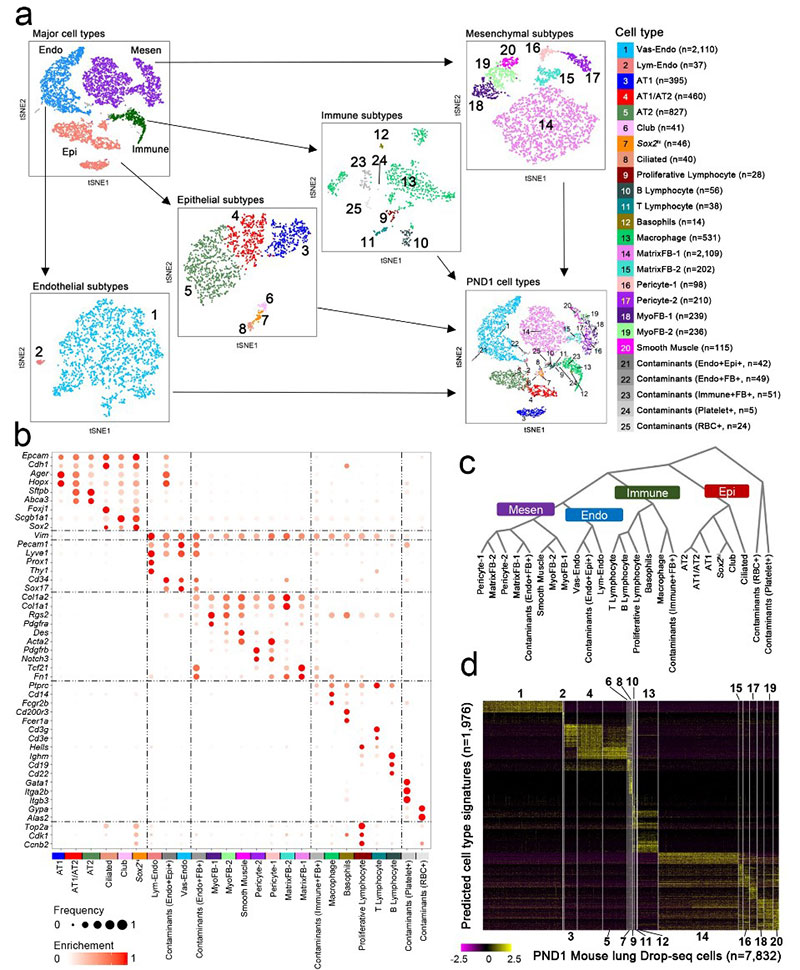
- Single cells RNA-seq from whole mouse lung at E16.5, E18.5, P1, P3, P7, P28 (Cincinnati Children's Hospital Medical Center)
- snRNA-seq & snATAC-seq of human lung from neonate, child and adult (UCSD, Cincinnati Children's & Univ. of Rochester)
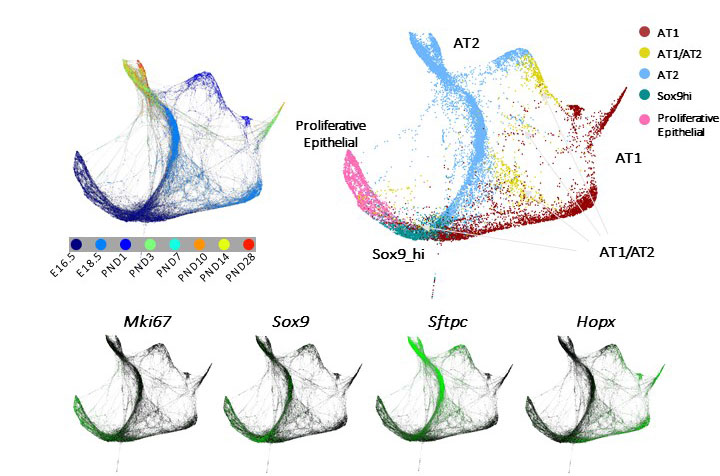
- Delineate lineage trajectory of the lung cell types
- Glucocorticoid Regulation of Pulmonary Mesenchymal Cell differentiation and lung maturation
- Unraveling transcriptome dynamics in lung development (E16.5-P28), characterize lineage dynamics of major lung cell type, identify niche, progenitor and differentiation states of individual cells
- Lung Diseases
- Single-cell RNA sequencing analysis of idiopathic pulmonary fibrosis lungs
- Single-cell RNA sequencing analysis of lungs with TBX4 mutation
- A single cell mapping of nasal epithelial cells from CF vs. non-CF patients, treated (CFTR modulators VX809/VX770) vs untreated.
- Single cell transcriptome analysis of airway submucosal glands of CF patients.
Grant Support
U01 HL148856 (Whitsett / Potter / Xu)
Chan Zuckerberg Foundation- HCA Lung Seed Network (Whitsett / Xu)
CFF Clinical Pilot and Feasibility Award (Xu)
P30 DK117467 (Naren, Whitsett) - Pilot and Feasibility (Xu)